Systematic drug repurposing to enable precision medicine: a case study in breast cancer
Precision medicine and drug repurposing provide an opportunity to ameliorate the challenges of declining pharmaceutical R&D productivity, rising costs of new drugs, and poor patient response rates to existing medications. Multifactorial “disease signatures” provide unique insights into the architecture of complex disease populations that can be used to better stratify patient groups, aiding the delivery of precision medicine.
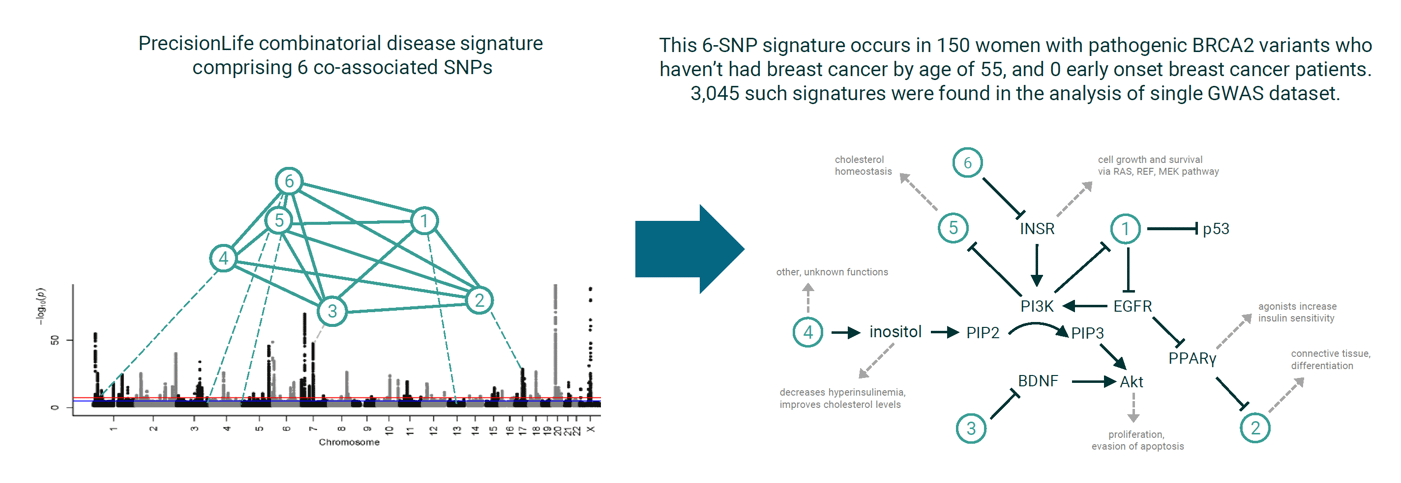
Analysis of a complex disease (breast cancer) population was undertaken to identify the combinations of single‑nucleotide polymorphisms that are associated with different disease subgroups. Target genes associated with the disease risk of these subgroups were examined, followed by identification and evaluation of existing active chemical leads as drug repurposing candidates.
One hundred and seventy‑five disease‑associated gene targets relevant to different subpopulations of breast cancer patients were identified. Twenty‑three of these genes were prioritized as both promising novel drug targets and repurposing candidates. Two targets, P4HA2 and TGM2, have high repurposing potential and a strong mechanistic link to breast cancer.
This study showed that detailed analysis of combinatorial genomic (and other) features can be used to accurately stratify patient populations and identify highly plausible drug repurposing candidates systematically across all disease‑associated targets.