Combinatorial Analytics & AI
Providing the most reliable, reproducible and detailed understanding of disease on the planet

Characterizing disease by molecular mechanism
Chronic diseases present the greatest challenge to human health. They're caused by a complex interplay of biology, which gold standard genomic analysis and AI models are unable to detect.
Hypothesis-free combinatorial analytics is uniquely able to reveal how multiple genes and other factors interact to cause complex diseases. Identifying the mechanisms specific to each patient to personalize care and improve health outcomes.
Our platform is unbiased and infinitely scalable, producing findings that are reproducible and explainable – making new discoveries faster than ever, with the highest level of confidence.
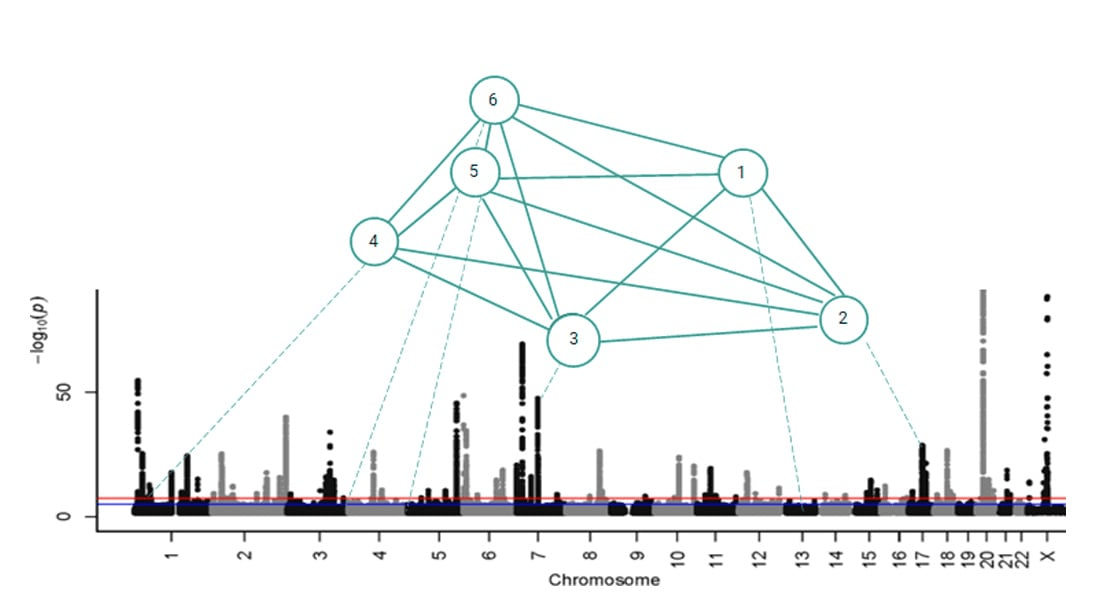
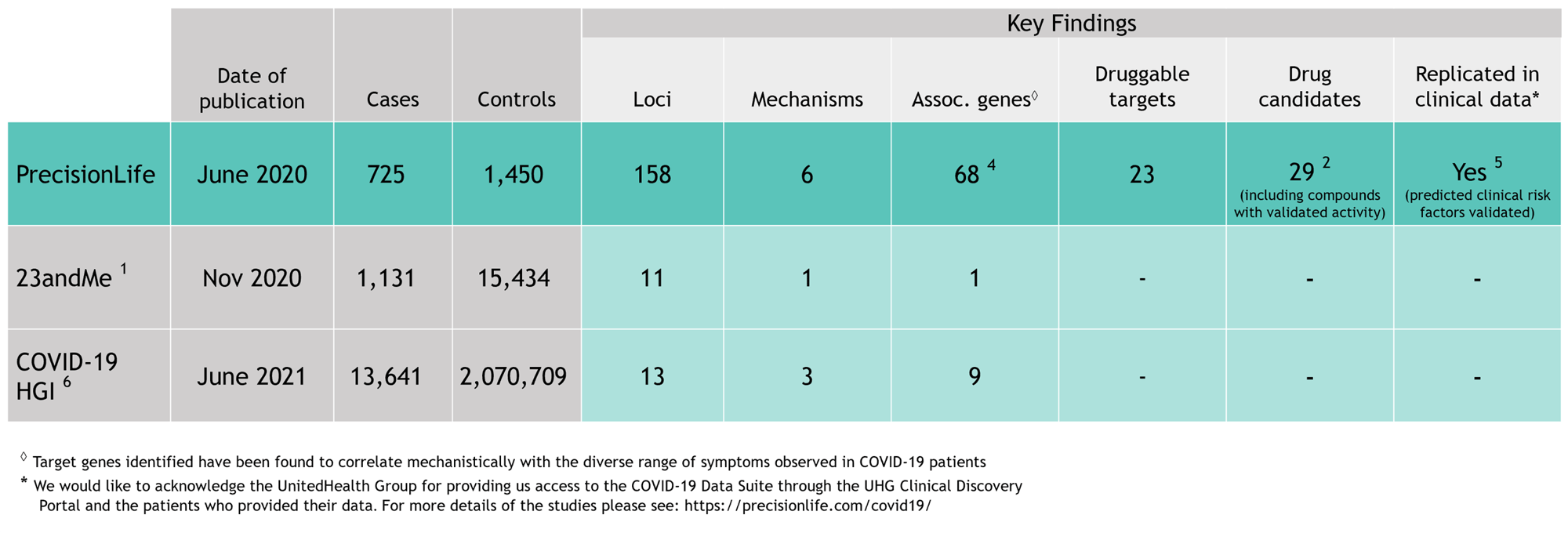
Combinatorial disease signature comprising six co-associated SNPs
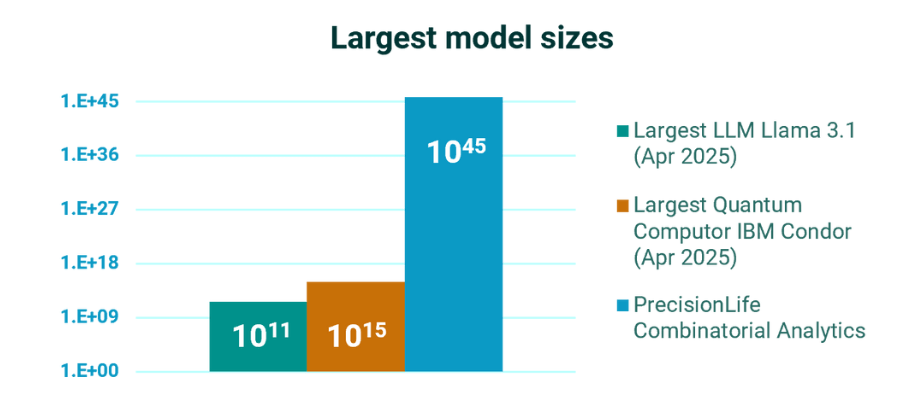
Going far beyond all other data models
AI models like LLMs are limited by their training sets, which are biased and too incomplete to address chronic disease biology.
Combinatorial analytics enables us to rapidly analyze multiomic datasets at massively higher orders of combinations – far more than is possible with even the most advanced data models.
We capture non-linear signal in the biology of genetic and metabolic networks – revealing the effects of combinations of features and the deepest insights into disease biology to de-risk every stage of drug development and clinical care.
Analyzing every combination to find the cause
Our platform rapidly analyzes the widest range of multimodal data types, singly or in combination, without bias to identify features that in combination are associated with a phenotype.
This allows us to generate much more and far richer signal from smaller datasets than traditional Genome Wide Association Studies (GWAS) models and to better stratify patients according to the mechanistic origins of their disease.
Multiomic
Genomic
Metabolomic
Proteomic
Transcriptomic
Phenotypic
Microbiome
Epidemiological
Clinical data
Imaging
Environmental
Pathogens
Structural variants
Digital biomarkers
Real world evidence
more
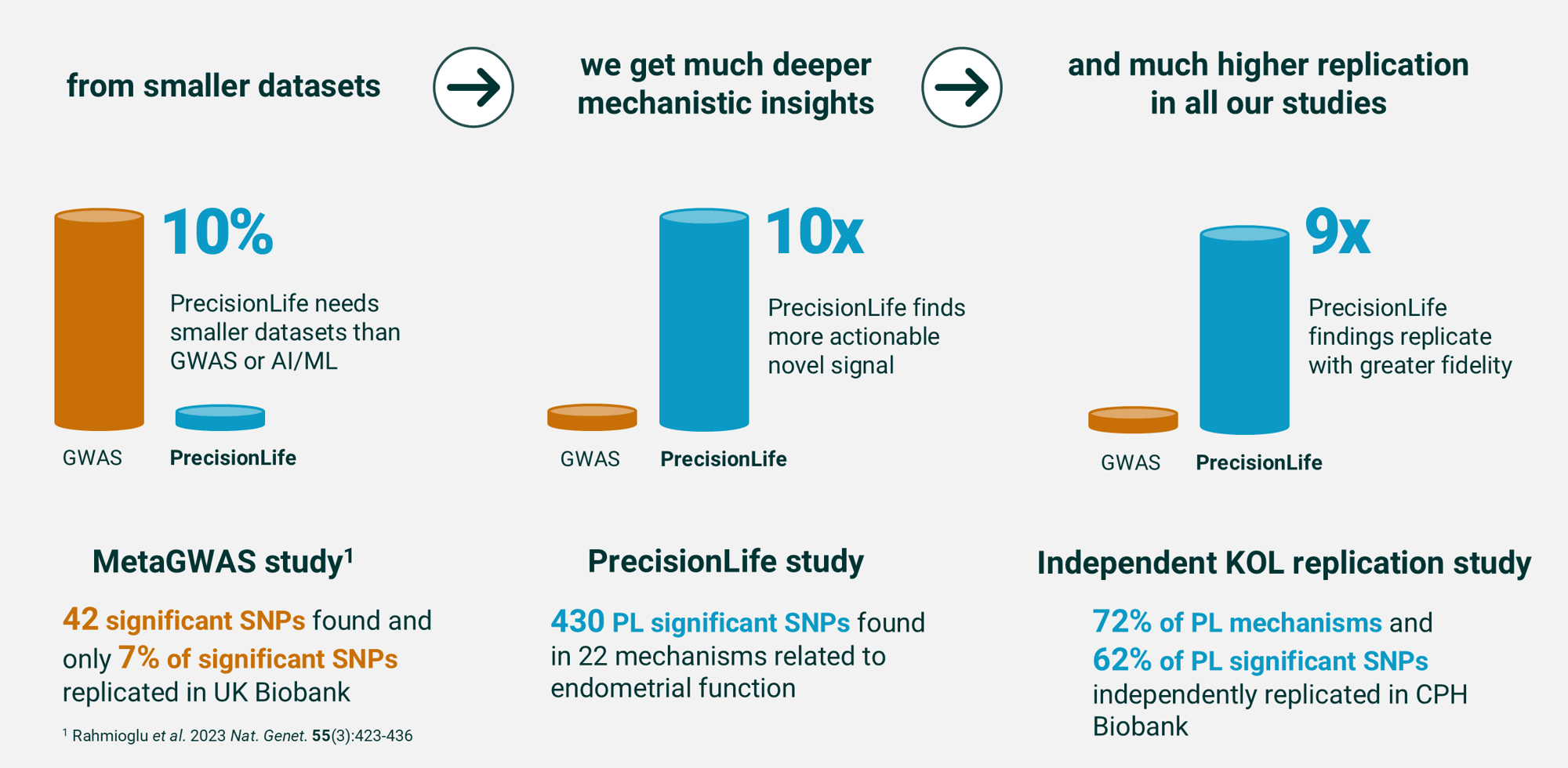
Greater understanding & higher replication
Combinatorial analytics identifies significantly more mechanistic insights, from less data, with considerably greater replication than any other analytical method.
The graphic below shows this in a direct comparison between the PrecisionLife platform and a meta-GWAS of endometriosis patient data, published in Nature Genetics¹ by an international consortium of key opinion leaders.
CASE STUDY: COVID-19
Deeper disease insights, faster
Our platform outperformed global efforts to identify the genetic factors of severe COVID-19 susceptibility, producing more insights, clinical biomarkers, and novel drug targets than any other analysis before or since.
Completed 12-months earlier than the COVID-19 Host Genetics Initiative (HGI), our analysis identified many more loci and more than 7X the number of novel genes associated with severe COVID-19 – compared to any other global research (as shown in the figure below) – and linked them all to major symptoms of the disease.
Our findings have since been extensively validated independently by research groups around the world.