Overview
The Global Initiative for Asthma estimates that 334 million people are affected by asthma. Patients that experience persistent symptoms, frequent asthma attacks, or low lung function despite taking asthma medication form a relatively large group of patients known as treatment-refractory asthmatics.
We analyzed SNP genotype data from 42,000 asthma patients using combinatorial analytics followed by k-means clustering and pathway enrichment to identify statistically significant combinations of up to five SNP genotypes.
Our analysis found that many of the most critical SNP clusters are related to immune system pathways, and antigen presentation and processing.
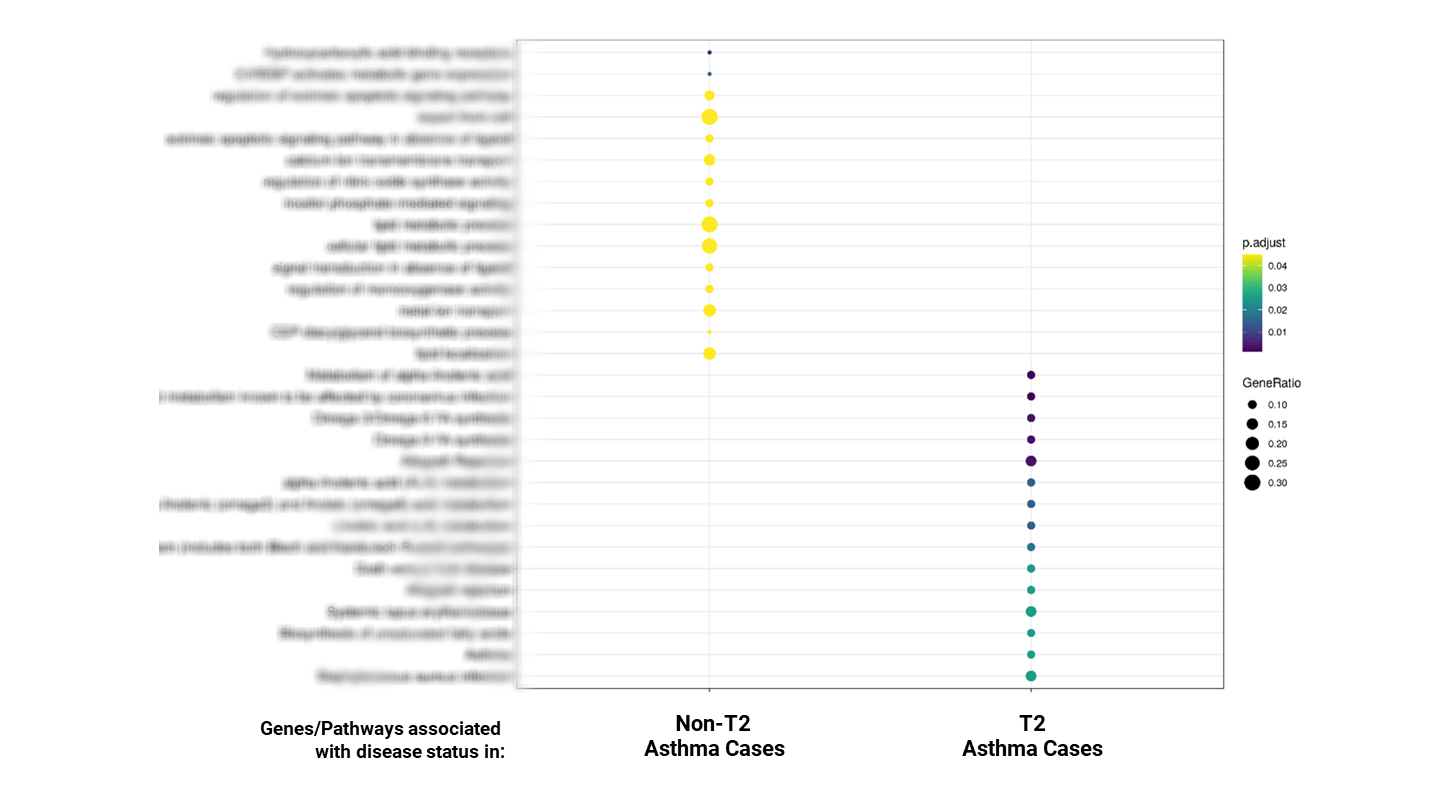
Our comparative study between the T2 and non-T2 asthma patient populations clearly demonstrated the opportunity and means to address the unmet medical need of non-T2 patients. These findings hold significant potential for better patient stratification, diagnosis biomarkers, and new treatment options.