Poster presented at Society for Neuroscience 2022
Alzheimer Disease Multiomics Session
Authors: K. Taylor, A. Malinowski, S. Das, S. Hogg
Download the poster
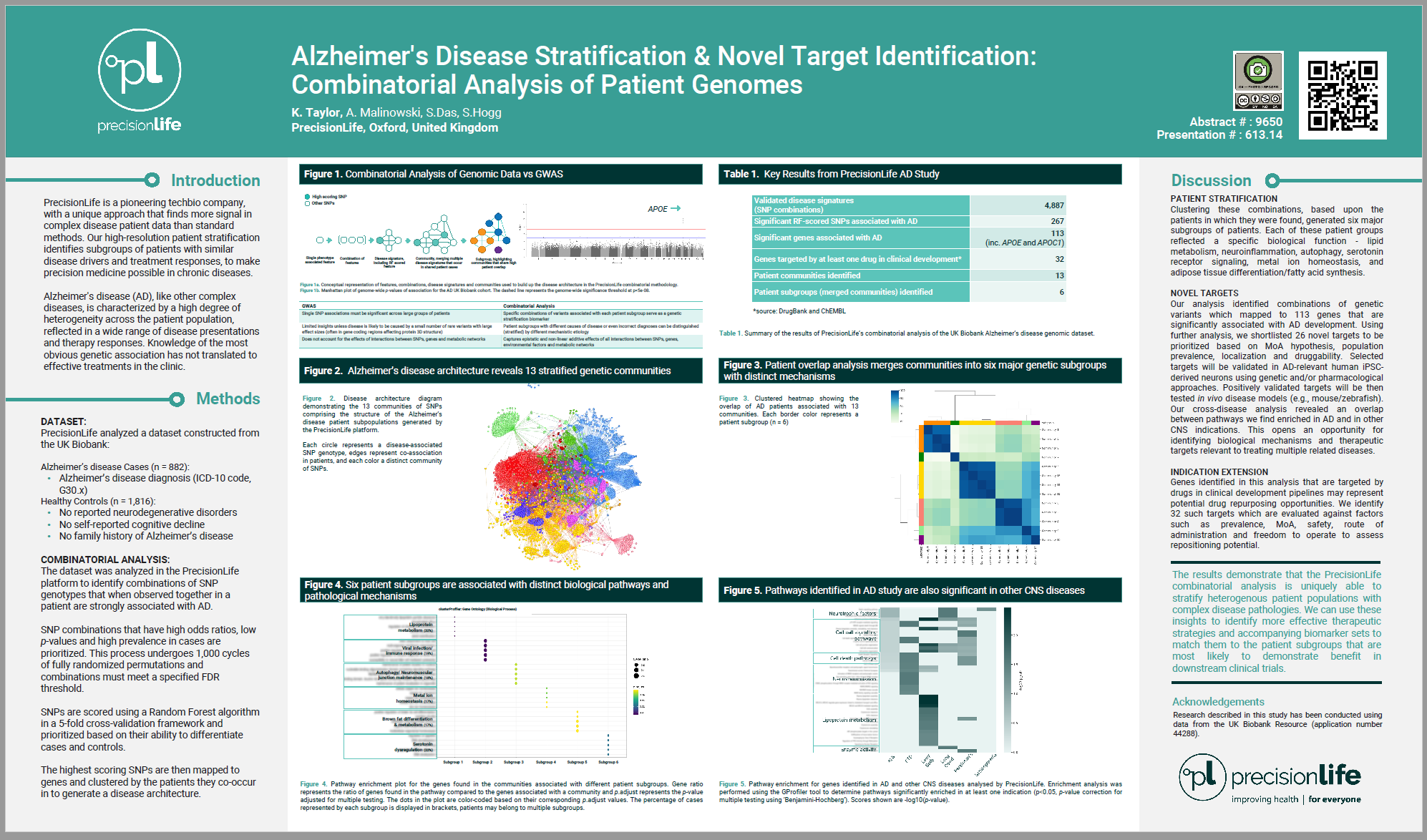
Alzheimer's disease (AD), like other complex diseases, is characterised by a high degree of heterogeneity across the patient population, reflected in a wide range of disease presentations and therapy responses. GWAS have identified several disease-associated genes, but these findings have not translated into progress in clinical trials. This probably reflects the fact that GWAS is limited to identifying single variants with large effect sizes in a population, while the key to understanding complex diseases such as AD that are influenced by multiple genetic loci, epidemiological and/or environmental factors is to find combinations of these disease associated factors that distinguish one patient subgroup from another.
The PrecisionLife platform utilises a hypothesis-free method for the detection of combinations of features that together are strongly associated with variations in disease risk, symptoms, progression rates and therapy response often observed in AD patient subgroups.
To explore the substructures within the AD population we analysed a genomic dataset from the UK Biobank, including 882 patients diagnosed with AD as defined by the ICD-10 code G30.x. These patients were compared against healthy controls with no reported neurodegenerative disorders, self-reported cognitive decline or AD, and no family history of AD. The oldest available controls (to minimize the potential for late onset disease) who met these criteria were gender-matched and selected in a 2:1 ratio against the cases.
Our analysis identified combinations of genetic variants which mapped to 113 genes that are significantly associated with AD development. Clustering these combinations, based upon the patients in which they were found, generated six major subgroups of patients. Each of these patient groups reflected a specific biological function - lipid metabolism, neuroinflammation, autophagy, serotonin receptor signaling, metal ion homeostasis, and adipose tissue differentiation/fatty acid synthesis. Furthermore, the specific combinations of variants associated with each patient subgroup serve as a biomarker to identify patients most likely to be responsive to pharmacological modulation of the target or pathways identified.
The results demonstrate that the PrecisionLife combinatorial analysis is uniquely able to stratify heterogenous patient populations with complex disease pathologies. We can use these insights to identify more effective therapeutic strategies and accompanying biomarker sets to match them to the patient subgroups that are most likely to demonstrate benefit in downstream clinical trials.
Alzheimer's disease study
Stratifying patient subgroups to reduce the risk of future clinical trial failure.
"This is a quote about something or other, test da ba dee da ba doo. Lorem Ispum Lorem Ipsum"
Name Surname, Position, Company