Overview
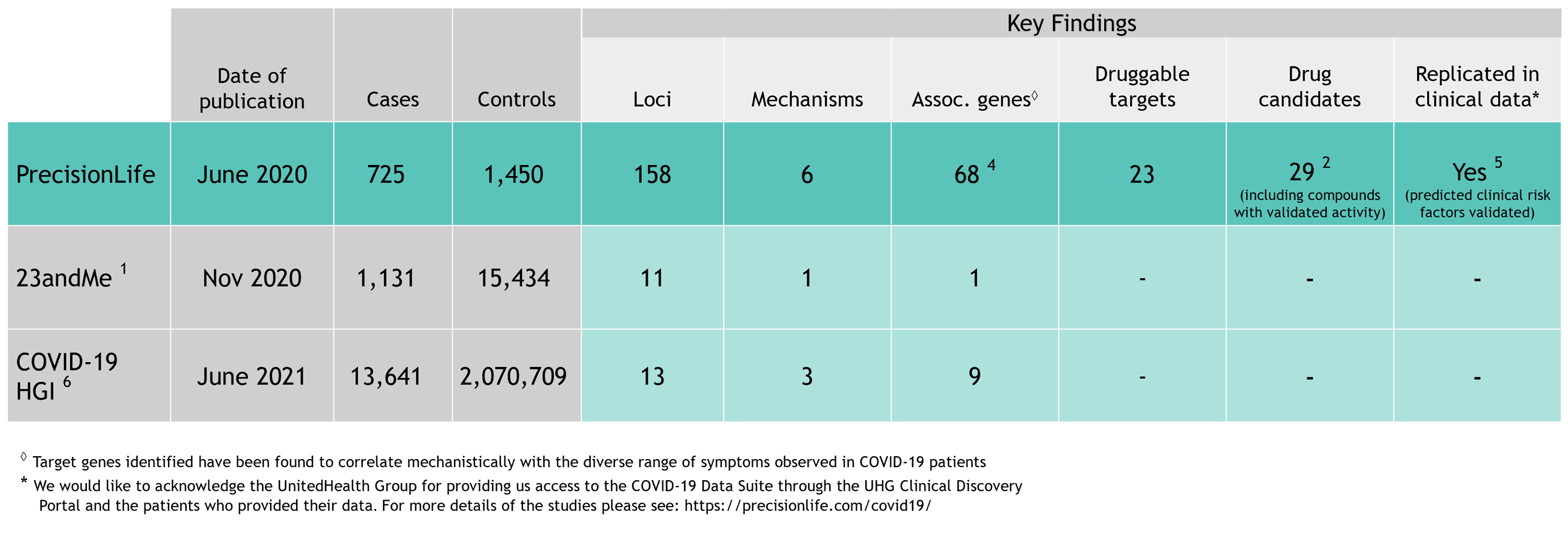
As the first wave of the COVID-19 pandemic surged around the world in early 2020, scientists and clinicians united in an unprecedented effort to understand the disease, its diverse symptoms and severities, and how to treat it. As part of this effort, we analyzed genetic host response risk factors in the first small cohort of 779 severe COVID-19 patients reported from the UK Biobank.
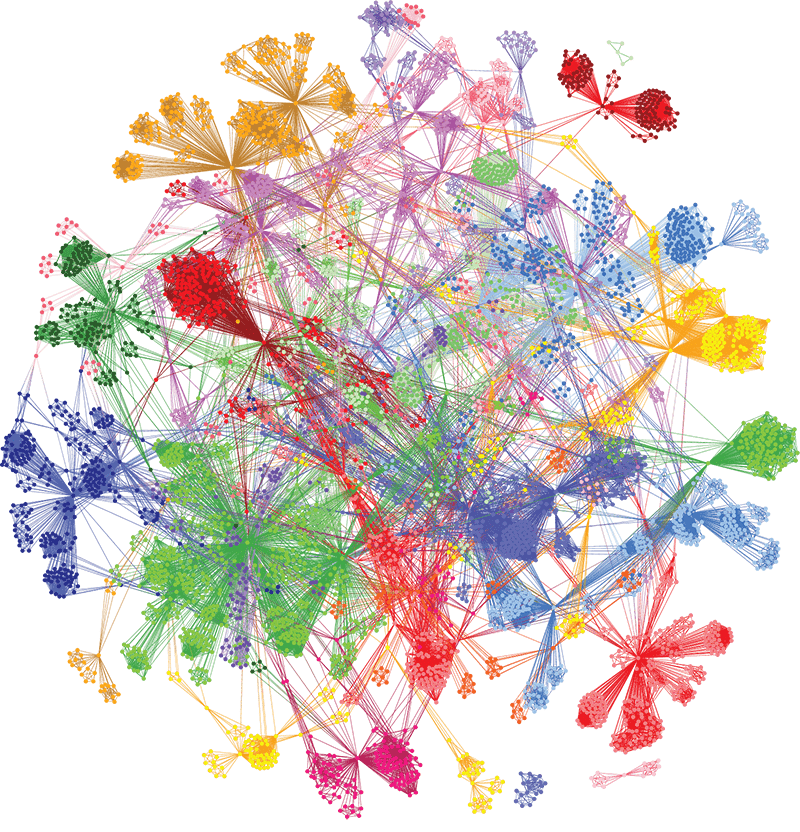
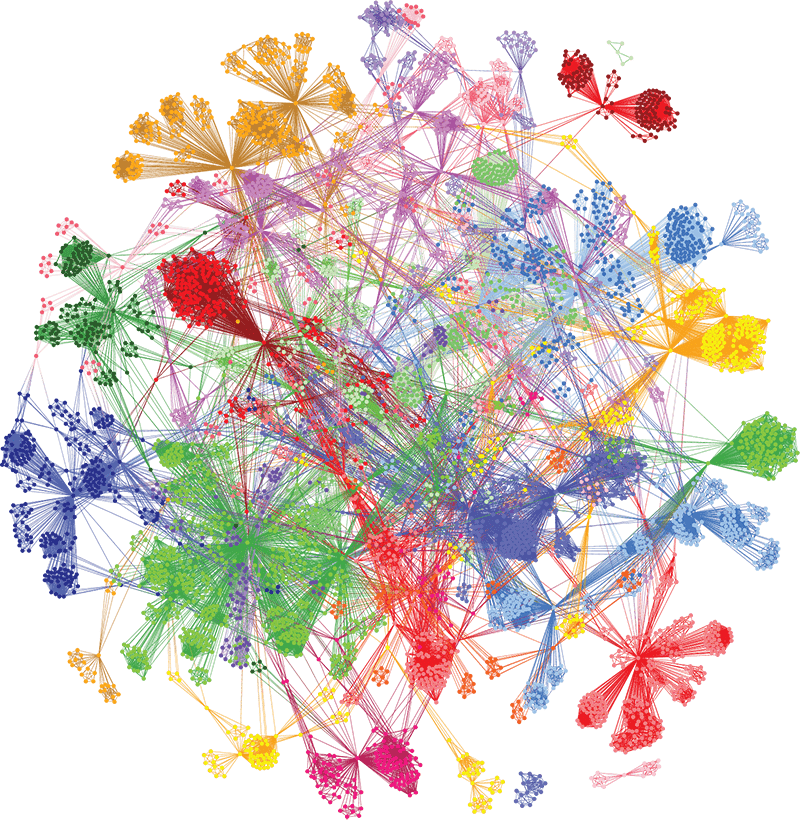
Using our unique combinatorial analytics platform, we identified drivers of different aspects of disease biology in specific patient subgroups and generated insights into the genetics, clinical presentation, pathology, and treatment of COVID-19 patients. The level of insights generated are not achievable with other analysis tools.